Difference between revisions of "201801 Pharmacogenomics CDS"
(Created page with "{{subst::Template for FHIR Connectathon Track Proposals}}") |
|||
| (20 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
[[Category:201801_FHIR_Connectathon_Track_Proposals|Jan 2018 Proposals]] | [[Category:201801_FHIR_Connectathon_Track_Proposals|Jan 2018 Proposals]] | ||
__NOTOC__ | __NOTOC__ | ||
| − | =Track | + | =Pharmacogenomics CDS= |
| + | |||
| + | |||
| + | *[https://drive.google.com/open?id=1sXe3sppykd8p2iHtc3GYKy4IH4BRWa5k Pharmacogenomics Track Orientation Slides] | ||
==Submitting WG/Project/Implementer Group== | ==Submitting WG/Project/Implementer Group== | ||
| − | + | Clinical Genomics WG | |
==Justification== | ==Justification== | ||
| Line 11: | Line 14: | ||
==Proposed Track Lead== | ==Proposed Track Lead== | ||
| − | + | Bob Dolin (BDolin@Elimu.io) | |
| + | |||
See [[Connectathon_Track_Lead_Responsibilities]] | See [[Connectathon_Track_Lead_Responsibilities]] | ||
==Expected participants== | ==Expected participants== | ||
| − | + | * Elimu | |
| + | * Cerner | ||
| + | * Children’s Mercy Hospital | ||
| + | |||
| + | Please contact track lead or post to CG list if you're interested in participating! | ||
==Roles== | ==Roles== | ||
| − | |||
<!-- Roles are sets of functionality (generally defined by a Conformance resource) that a single system can take on --> | <!-- Roles are sets of functionality (generally defined by a Conformance resource) that a single system can take on --> | ||
| − | === | + | ===PGx CDS Service=== |
| − | + | PGx CDS Service will interact with EHR (via CDS Hooks), OMIC Data Store (for genomoic observations), and Terminology Server (for medication value sets). | |
| + | |||
| + | ===OMIC Data Store=== | ||
| + | Will store genomic observations | ||
| + | |||
| + | ===EHR=== | ||
| + | Order entry of a medication triggers a “medication-prescribe” CDS hook which invokes the PGx CDS Service and provides it with a FHIR MedicationOrder instance. | ||
| + | |||
| + | ===Terminology Service=== | ||
| + | The PGx CDS Service will ping the terminology server to see if the ordered drug is in an “Actionable PGx Drug” value set. | ||
==Scenarios== | ==Scenarios== | ||
| − | |||
| − | + | Please see detailed storyboard here: https://docs.google.com/document/d/1VqPrf6aaOB8RhSbAq2JYDxZYbDm-RT9DxA0u6-RxYo0/edit | |
| − | : | ||
| − | |||
| − | |||
| − | |||
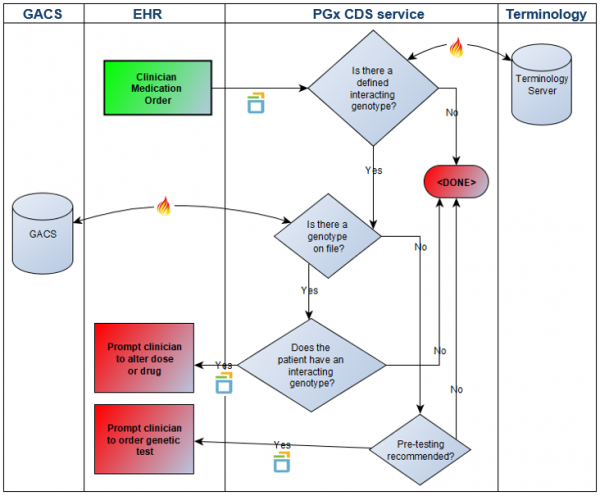
| − | + | A swim-lane overview of the scenario is here: | |
| + | |||
| + | [[File:PharmacogenomicsCDS.png|600px]] | ||
| + | |||
| + | |||
| + | :'''Action''': [1] (EHR) Manually enter an order for "Imuran 50mg 1 tablet twice a day by mouth" into order entry system. This triggers a “medication-prescribe” CDS hook with a hook context of MedicationOrder which includes RxNorm code 197388 for azathioprine 50mg oral tablet; [2] (PGx CDS Service) Having been triggered by the “medication-prescribe” CDS hook, the PGx CDS Service now executes a decision support rule that [1] determines if the ordered drug has a known gene interaction; [2] determines, where there is a known drug-gene interaction, whether or not the patient has genetic test results on file; [3] determines, where there are genetic test results on file, if the patient has an interacting genotype; [4] determines, where there are not genetic test results on file, if the patient needs pre-testing. PGx CDS Service returns a CDS hooks “information card” back to the EHR, with appropriate recommendations. | ||
| + | |||
| + | :'''Precondition''': Genomic observation(s) are available for some patients. | ||
| + | |||
| + | :'''Success Criteria''': Retrieve relevant observations (on TPMT gene, on allelic state) where they exist. | ||
| + | |||
| + | :'''Bonus point''': Search OMIC data store for different potential representations. | ||
| + | |||
| + | :'''Reference''': Azathioprine Pharmacogenomics - CPIC recommendations [https://cpicpgx.org/guidelines/guideline-for-thiopurines-and-tpmt/] | ||
==TestScript(s)== | ==TestScript(s)== | ||
| Line 48: | Line 72: | ||
I am happy to help: JohnMoehrke@gmail.com -- security co-chair | I am happy to help: JohnMoehrke@gmail.com -- security co-chair | ||
--> | --> | ||
| + | |||
| + | ==Findings and Conclusions== | ||
| + | Findings and conclusions from the session are captured here: | ||
| + | http://wiki.hl7.org/index.php?title=Record_Connectathon_17_Outcomes_Here#Pharmacogenomics_CDS | ||
Latest revision as of 20:42, 4 April 2018
Pharmacogenomics CDS
Submitting WG/Project/Implementer Group
Clinical Genomics WG
Justification
Proposed Track Lead
Bob Dolin (BDolin@Elimu.io)
See Connectathon_Track_Lead_Responsibilities
Expected participants
- Elimu
- Cerner
- Children’s Mercy Hospital
Please contact track lead or post to CG list if you're interested in participating!
Roles
PGx CDS Service
PGx CDS Service will interact with EHR (via CDS Hooks), OMIC Data Store (for genomoic observations), and Terminology Server (for medication value sets).
OMIC Data Store
Will store genomic observations
EHR
Order entry of a medication triggers a “medication-prescribe” CDS hook which invokes the PGx CDS Service and provides it with a FHIR MedicationOrder instance.
Terminology Service
The PGx CDS Service will ping the terminology server to see if the ordered drug is in an “Actionable PGx Drug” value set.
Scenarios
Please see detailed storyboard here: https://docs.google.com/document/d/1VqPrf6aaOB8RhSbAq2JYDxZYbDm-RT9DxA0u6-RxYo0/edit
A swim-lane overview of the scenario is here:
- Action: [1] (EHR) Manually enter an order for "Imuran 50mg 1 tablet twice a day by mouth" into order entry system. This triggers a “medication-prescribe” CDS hook with a hook context of MedicationOrder which includes RxNorm code 197388 for azathioprine 50mg oral tablet; [2] (PGx CDS Service) Having been triggered by the “medication-prescribe” CDS hook, the PGx CDS Service now executes a decision support rule that [1] determines if the ordered drug has a known gene interaction; [2] determines, where there is a known drug-gene interaction, whether or not the patient has genetic test results on file; [3] determines, where there are genetic test results on file, if the patient has an interacting genotype; [4] determines, where there are not genetic test results on file, if the patient needs pre-testing. PGx CDS Service returns a CDS hooks “information card” back to the EHR, with appropriate recommendations.
- Precondition: Genomic observation(s) are available for some patients.
- Success Criteria: Retrieve relevant observations (on TPMT gene, on allelic state) where they exist.
- Bonus point: Search OMIC data store for different potential representations.
- Reference: Azathioprine Pharmacogenomics - CPIC recommendations [1]
TestScript(s)
Security and Privacy Considerations
Findings and Conclusions
Findings and conclusions from the session are captured here: http://wiki.hl7.org/index.php?title=Record_Connectathon_17_Outcomes_Here#Pharmacogenomics_CDS