This wiki has undergone a migration to Confluence found Here
Specimen
Back to Orders & Observations WG main page
Back to OO Projects page
Contents
Scope of Specimen Project
The scope of the project is to:
- Complete DAM incorporating various use cases (OO, CG, II, AP)
- Complete Specimen V3 models based on other use cases (CG, II, AP)
- Consider V2 Updates
- Support Specimen Identifier formats as further established by AP workgroup
Leadership
Topic leads:
- Review/refine DAM – Joyce Hernandez, Mukesh Sharma
- Starting point: NCI Life Sciences DAM: Specimen Core.
- V3 CMET/RMIM compare + updates – Lorraine Constable
- V2 SPM compare + updates – Riki Merrick, Hans Buitendijk
- Validate Specimen Identifier format support - TBD
Conference Calls and Minutes
- Dates: Every other Thursday, starting Feb 16, 2012
- Time: 12:00 - 13:00 EST
- Phone: +1 770-657-9270
- Passcode: 653212
- Webex: http://www.mymeetings.com/siemens/nc/join.php?i=741301183&p=&t=c
- Passcode: HL7OO
- List Serve: specimen@hl7.lists.org
Minutes
- File:SPM Minutes 20120216 ConCall.doc
- File:SPM Minutes 20120329 ConCall.doc
- File:SPM Minutes 20120412 ConCall.docx
- File:SPM Minutes 20120524 ConCall.doc
- File:SPM Minutes 20120621 ConCall.docx
- File:SPM Minutes 20120705 ConCall.docx
- File:SPM Minutes 20120816 ConCall.docx
- File:SPM Minutes 20121018 ConCall.docx
- File:SPM Minutes 20121101 ConCall.docx
- File:SPM Minutes 20121115 ConCall.docx
- File:SPM Minutes 20121213 ConCall.docx
- File:SPM Minutes 20130110 ConCall.docx
- File:SPM Minutes 20130131 ConCall.docx
- File:SPM Minutes 20130214 ConCall.docx
- File:SPM Minutes 20130307 ConCall.docx
- File:SPM Minutes 20130314 ConCall.docx
- File:SPM Minutes 20130328 ConCall.docx
- File:SPM Minutes 20130605 ConCall.docx
- Minutes 20131114 ConfCall
Working Documents
Use Cases
- Clinical Sequencing Project - Identified Use Cases to Consider in Specimen CMET - from CG ClinSeq.doc
- Specimen Use Case for Isolate Representation
- Medical Research Use Case and Specimen Core Diagram Media:Specimen-Core Model Diagram and Medical Research Use Case Process Flow.xls
- Origin of Specimen
- Need to indicate whether the specimen is human, animal, soil, etc.
- For viral genetics where DNA/RNA can be extracted from a micro-organism.
- Notes from May WGM
- II perspective mainly interested in specimen in context of interventional imaging
- An introduction to the DICOM specimen information model is available in DICOM Part 17 (available free at ftp://medical.nema.org/medical/dicom/2011/11_17pu.pdf) Annex NN.
- Detailed specimen information can be found in DICOM Part 3 (available free at ftp://medical.nema.org/medical/dicom/2011/11_03pu.pdf). This information that has been specified by DICOM WG26 "Pathology" (DICOM Supplement 122 "Specimen Identification and Revised Pathology" Project) should be taken into consideration in the project.
Status & Deliverables
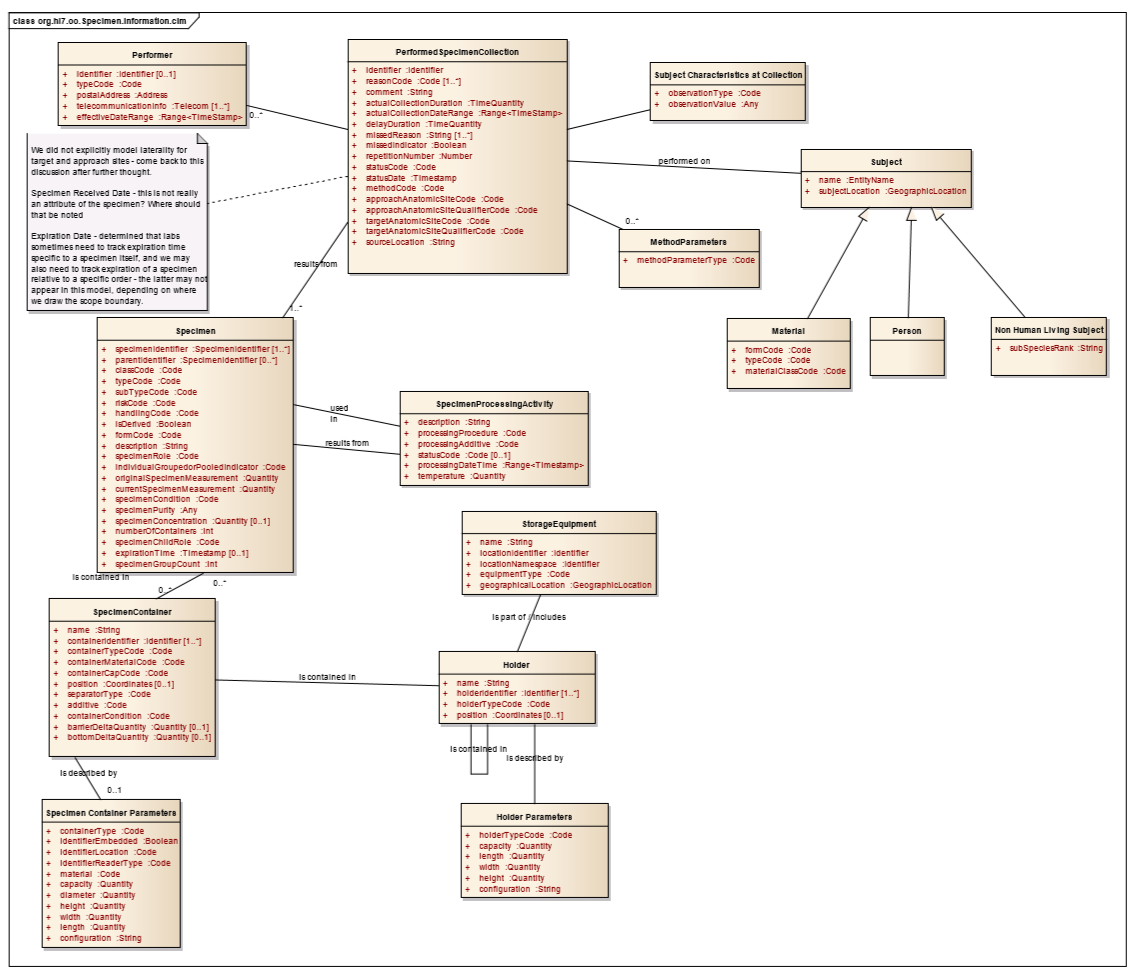
Draft Model
Specimen Model in Gforge: http://gforge.hl7.org/gf/project/orders/scmsvn/?action=browse&path=%2Ftrunk%2FPOSP%2FDAM%2F
Definitions spreadsheet: File:Specimen DAM classes and attributes with notes.xlsx
- snapshot of Draft Model work in progress (draft model on the left, excerpts from NCI LSDAM on the right):
Links
LS DAM Wiki: https://wiki.nci.nih.gov/x/cxRlAQ
LS DAM Overview: https://wiki.nci.nih.gov/download/attachments/76677702/LSDAMSpecimenHL7OO.pptx
LS DAM (Enterprise Architect) : https://wiki.nci.nih.gov/download/attachments/23401587/LSDAMWorking.EAP
LS DAM (HTML): http://ncientarch.info/sandbox/LSDAM2_2_2/EARoot/EA3/EA1/EA1/EA1/EA1/EA1/EA4/EA157.htm